- Accueil
- Volume 19 (2015)
- Numéro 2
- New lipolytic enzymes identified by screening two metagenomic libraries derived from the soil of a winter wheat field
Visualisation(s): 1168 (9 ULiège)
Téléchargement(s): 57 (1 ULiège)
New lipolytic enzymes identified by screening two metagenomic libraries derived from the soil of a winter wheat field

Notes de la rédaction
Received on August 21, 2014; accepted on January 9, 2015
Résumé
Nouvelles enzymes lipolytiques identifiées par criblage de deux banques métagénomiques construites à partir du sol d’un champ de blé d’hiver
Description du sujet. Les enzymes lipolytiques sont largement distribuées dans différents environnements et remplissent des rôles physiologiques importants au sein des micro-organismes qui les peuplent. Les sols représentent des environnements riches et diversifiés contenant des communautés microbiennes largement méconnues.
Objectifs. Ce travail a pour but de découvrir de nouvelles enzymes lipolytiques.
Méthode. Les nouvelles enzymes ont été isolées par criblage fonctionnel de deux banques métagénomiques (une banque d’hiver et une banque de printemps) construites à partir d’un sol agricole. Les criblages ont été réalisés sur milieu 2xYT contenant 3 % de réactif pour lipase.
Résultats. Dix-neuf candidats positifs ont été isolés. L’analyse des inserts de ces candidats a mené à l’identification de 23 enzymes lipolytiques potentielles (13 dans la banque d’hiver et 10 dans celle de printemps). Ces enzymes présentent entre 31 % et 62 % d’identité avec des enzymes connues et appartiennent à sept familles différentes.
Conclusions. Comme ces enzymes démontrent une faible identité avec des enzymes connues, elles peuvent présenter de nouvelles caractéristiques biochimiques.
Abstract
Description of the subject. Lipolytic enzymes are widely distributed and fulfil important physiological functions in the microorganisms inhabiting diverse environments. Soils are rich, diversified environments containing microbial communities that remain largely unknown.
Objectives. This work aimed to discover new lipolytic enzymes.
Method. New enzymes were found by functional screening of two seasonal metagenomic libraries (a winter and a spring library) constructed from an agricultural soil. Screens were performed on 2xYT medium supplemented with 3% lipase reagent.
Results. Nineteen positive clones were isolated. Analysis of the corresponding inserts led to identifying 23 putative lipolytic enzymes (13 for the winter library and 10 for the spring library) displaying between 31% and 62% identity to known enzymes and belonging to seven different families.
Conclusions. As enzymes show low identity to known enzymes, the encoded enzymes may display novel biochemical features.
Table des matières
1. Introduction
1Carboxylic ester hydrolases (EC 3.1.1.x) belonging to the α/β hydrolase superfamily are widely distributed and perform important physiological functions in the bacteria, archaea, fungi and plants inhabiting diverse environments. They are among the most important biocatalysts for biotechnological applications in agriculture, food or pharmaceutical industries for example (Panda et al., 2005; Casas-Godoy et al., 2012; Fazary et al., 2013). They include carboxylesterases (EC 3.1.1.1), hydrolyzing (at least partly) water-soluble short-acyl-chain esters, and triacylglycerol lipases (EC 3.1.1.3), preferring water-insoluble long-chain triglycerides (Arpigny et al., 1999). The diverse origins and substrate specificities of lipolytic enzymes make it hard to classify them. Several classifications exist (Lenfant et al., 2013). The most employed is that of Arpigny and Jaeger which currently distinguishes 16 lipolytic enzyme families (I to XVI) on the basis of amino-acid sequence and fundamental biological properties (Arpigny et al., 1999; Handrick et al., 2001; Ewis et al., 2004; Lee et al., 2006; Levisson et al., 2007; Kim et al., 2009; Rao et al., 2011; Charbonneau et al., 2013; Fu et al., 2013). The ESTHER database, which classifies the α/β-hydrolase-fold proteins, has integrated under new names all the families of Arpigny and Jaeger except families II and VIII, belonging to the SGNH-hydrolase and the β-lactamase fold (Lenfant et al., 2013). Additionally, carbohydrate esterases are classified in the Carbohydrate-Active Enzymes Database (Cantarel et al., 2009) and thioesterases, in the Thioester-Active Enzyme Database (Cantu et al., 2011).
2Soils represent a major source of enzymes and other biomolecules of industrial importance (Strohl, 2000). Here we aimed to isolate new lipolytic enzymes from a microbiologically very diversified agricultural soil (Stroobants et al., 2014a), which has already yielded several β-glycosidases (Stroobants et al., 2014b). Screening of two metagenomic libraries (a winter and a spring library) has led to identifying 23 putative esterases belonging to seven different families.
2. Materials and methods
2.1. Soil description, sampling and library construction
3Soil samples were taken from a well-characterized experimental field located in Gembloux (50°33’N, 4°42’E), Belgium. The soil has been described previously (Stroobants et al., 2014a), as have the soil sampling and library construction procedures (Biver et al., 2014; Stroobants et al., 2014b).
2.2. Library screening and sequence analyses
4After pooling recombinants obtained for each library as explained in Stroobants et al. (2014b), about 210,000 Escherichia coli colonies per library (~1,100 Mb/library) were screened for lipolytic activity by plating the transformants on 2xYT medium (MP Biomedicals, Solon, OH, USA) containing 15 g·l-1 agar (Oxoid Ltd., Basingstoke, Hampshire, UK), 50 µg·ml-1 ampicillin and 30 g·l-1 lipase reagent (Difco, Detroit, MI, USA), a mixture of tributyrin and polysorbate 80. The plates were incubated at 37 °C for two days and then transferred to room temperature (~22 °C) for two to three weeks. Lipolytic activity was detected by observation of a halo surrounding each positive colony and was confirmed by isolating the plasmids of positive clones and introducing them into “Subcloning Efficiency” chemically competent DH5α E. coli cells (Invitrogen, Carlsbad, CA) before spreading onto the same medium as used for screening. After confirmation, the inserts were sequenced at GATC Biotech (Konstanz, Germany) and the BLASTX program was used to compare the insert sequences with Genbank sequences. Open reading frames (ORFs) were sought with the NCBI ORF Finder. Signal peptide cleavage sites were predicted with the SignalP 4.0 server (Petersen et al., 2011). Theoretical isoelectric points and molecular weights were computed with the Compute pI/Mw tool of ExPASy (Artimo et al., 2012). Conserved domains were identified with the NCBI CD-Search software (Marchler-Bauer et al., 2011). The Blast tool of the ESTHER database was used to assign putative α/β hydrolases to ESTHER families (Lenfant et al., 2013). Arpigny and Jaeger families were determined by amino acid sequence alignment (data not shown).
2.3. Sequence accession numbers
5The insert sequences of the positive candidates have been deposited in the GenBank database under accession numbers KM359523 to KM359541.
3. Results and discussion
3.1. Library construction and screening
6A winter and a spring library, each containing approximately 100,000 DH10B E. coli clones, were constructed from metagenomic DNA extracted from agricultural soil samples. Approximately 98% of the library clones had inserts ranging in size from 3 to 11 kb, with an average of 5.5 kb. About 210,000 colonies per library were screened for lipolytic activity on 2xYT agar medium supplemented with lipase reagent. Eleven candidates were found in the winter library and eight in the spring library. These clones were given names based on the model “cl-(W/S)-AS-TribX”, where X is a one- or two-digit number and W and S are used to differentiate clones isolated from winter or spring library. In what follows, a clone name without the “cl” designates either an ORF (italics) or a predicted protein (no italics).
3.2. Sequence analysis of positive candidate inserts and lipolytic activity of (sub)clones
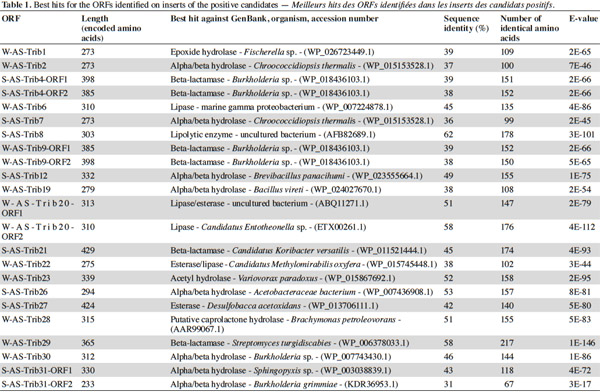
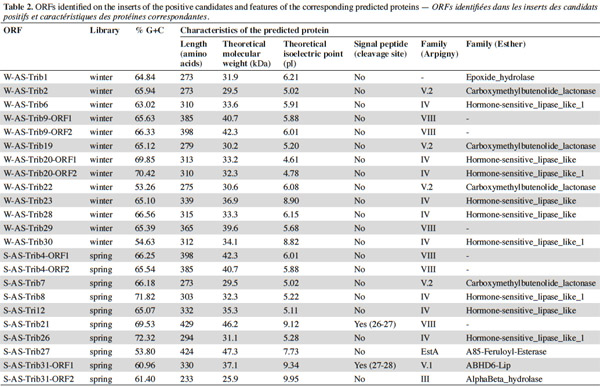
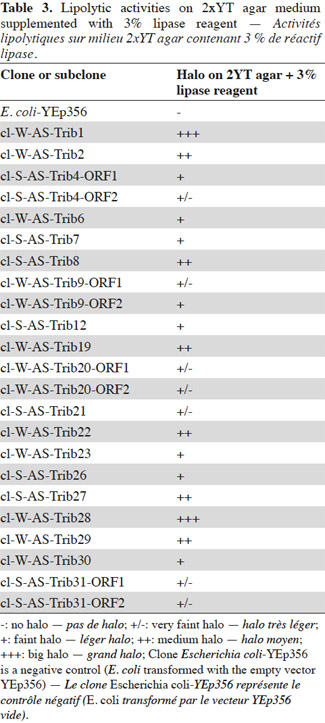
7After extraction, the plasmid inserts of the positive candidates (3,890 to 9,813 bp long) were sequenced and analyzed with the BLASTX program and ORF Finder. The identified ORFs and predicted proteins are shown in tables 1 and 2. The putative lipolytic enzymes showed only 31% to 62% identity to known ones, in agreement with the fact that at a depth of 10-20 cm, unknown species make up half of the bacterial community of the soil sampled in this study (Stroobants et al., 2014a) and in accordance with other soil screenings (Nacke et al., 2011; Biver et al., 2013). Four inserts displayed two ORFs coding for putative lipolytic enzymes (corresponding subclones: cl-W/S-AS-TribX-ORF1 and cl-W/S-AS-TribX-ORF2). The lipolytic activity of the (sub)clones was estimated from the size of the clear halo observed after plating on lipase-reagent-containing medium (Table 3). The activity observed was high for two, medium for six, low for eight, and very low for seven clones.
3.3. Classification of the putative lipolytic enzymes
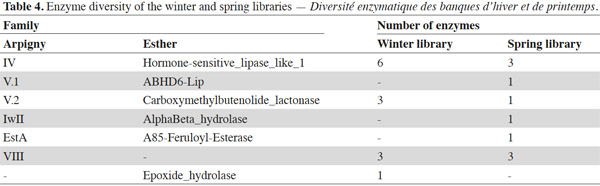
8To classify the putative lipolytic enzymes, we used the two main classification systems: Arpigny and Jaeger (Arpigny et al., 1999) and ESTHER (for α/β hydrolases) (Table 2). All but six of the predicted proteins belong to the α/β hydrolase superfamily. The six exceptions, showing striking similarity to several β-lactamases and containing the pfam domain 00144 typical of β-lactamases, were assigned to Arpigny and Jaeger’s family VIII. As observed in many other metagenomic studies of soil samples (Lee et al., 2004; Hong et al., 2007; Nacke et al., 2011), Arpigny and Jaeger’s family IV (called the Hormone-sensitive lipase-like family in ESTHER) was found to preponderate, with six representatives for the winter library and three for the spring library, all sharing the conserved motifs typical of this family: HGGG and GDSAG (where the serine is involved in the catalytic triad) (Arpigny et al., 1999). S-AS-Trib31-ORF2, assigned to family III, contains the conserved GxS(L/M)GG motif typical of this family, preceded by the conserved amino acids PG. S-AS-Trib27, which possesses the GHSMG pentapeptide conserved in the family III-related EstA-family esterases (Chu et al., 2008), was assigned to the EstA family. Six putative lipolytic enzymes could not be clearly classified in the Arpigny and Jaeger system, so the ESTHER system was used. Four predicted proteins were thus assigned to the Carboxymethylbutenolide lactonase family (family V.2 of Arpigny and Jaeger) (Lenfant et al., 2013), characterized by the presence of the conserved GxSxG and PT motifs typical of family V in addition to the conserved 3-oxoadipate enol-lactonase domain (TIGR02427). This domain is present also in W-AS-Trib1, assigned to the Epoxide hydrolase family, which accordingly includes some carboxymethylbutenolide lactonases. S-AS-Trib31-ORF1, assigned to the ABHD6-Lip family (family V.1 of Arpigny and Jaeger), possesses the GxSxG motif in addition to the the SxGG and D(L/M)PG motifs of this family.
9Some aspects of the observed distribution of ORFs between the winter and spring libraries seem worth noting (Table 4). For example, the spring library seemed more diverse, even though, lipolytic-enzyme-encoding ORFs seemed more abundant in the winter library (6 [sub]families for 10 ORFs versus 4 families for 13 ORFs). On the other hand, a high similarity was observed between certain winter-library and some spring-library predicted proteins: W-AS-Trib9-ORF1 and W-AS-Trib9-ORF2 are approximately 96% identical to S-AS-Trib4-ORF1 and S-As-Trib4-ORF2; W-AS-Trib2 is 97% identical to S-AS-Trib7. This suggests some similarity at the level of the bacterial species composing the winter and spring microbial communities of this soil.
4. Conclusions
10By screening two metagenomic libraries obtained from an agricultural soil, we have identified sequences encoding 23 putative lipolytic enzymes belonging to 7 families and displaying 31% to 62% identity to known lipolytic enzymes. As they show low identity to known enzymes, the encoded enzymes may display novel biochemical features. This could be of interest to industrialists on the lookout for novel lipolytic enzymes for use in agriculture or the production of foods or pharmaceuticals. The next step will thus be to produce and characterize the corresponding proteins.
Bibliographie
Arpigny J.L. & Jaeger K.E., 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J., 343(1), 177-183.
Artimo P. et al., 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res., 40(W1), W597-603.
Biver S. & Vandenbol M., 2013. Characterization of three new carboxylic ester hydrolases isolated by functional screening of a forest soil metagenomic library. J. Ind. Microbiol. Biotechnol., 40(2), 191-200.
Biver S. et al., 2014. Two promising alkaline β-glucosidases isolated by functional metagenomics from agricultural soil, including one showing high tolerance towards harsh detergents, oxidants and glucose. J. Ind. Microbiol. Biotechnol., 41(3), 479-488.
Cantarel B.L. et al., 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res., 37, D233-238.
Cantu D.C. et al., 2011. ThYme: a database for thioester-active enzymes. Nucleic Acids Res., 39, D342-346.
Casas-Godoy L. et al., 2012. Lipases: an overview. Methods Mol. Biol., 861, 3-30.
Charbonneau D.M. & Beauregard M., 2013. Role of key salt bridges in thermostability of G. thermodenitrificans EstGtA2: distinctive patterns within the new bacterial lipolytic enzyme family XV. PLoS One, 8(10), e76675, doi: 10.1371/journal.pone.0076675.
Chu X. et al., 2008. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl. Microbiol. Biotechnol., 80(4), 615-625.
EwisH.E. et al., 2004. Molecular cloning and characterization of two thermostable carboxyl esterases from Geobacillus stearothermophilus. Gene, 329, 187-195.
Fazary A.E. & Ju Y.-H., 2013. The large-scale use of feruloyl esterases in industry. Biotechnol. Mol. Biol. Rev., 3(5), 95-110.
Fu J. et al., 2013. Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol., 97(9), 3965-3978.
Handrick R. et al., 2001. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J. Biol. Chem., 276(39), 36215-36224.
Hong K.S. et al., 2007. Selection and characterization of forest soil metagenome genes encoding lipolytic enzymes. J. Microbiol. Biotechnol., 17(10), 1655-1660.
Kim E.Y. et al., 2009. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl. Environ. Microbiol., 75, 257-260.
Lee M.H. et al., 2006. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl. Environ. Microbiol., 72, 7406-7409.
Lee S.W. et al., 2004. Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl. Microbiol. Biotechnol., 65(6), 720-726.
Lenfant N. et al., 2013. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res., 41, D423-429.
Levisson M. et al., 2007. Characterization and structural modeling of a new type of thermostable esterase from Thermotoga maritima. FEBS J., 274(11), 2832-2842.
Marchler-Bauer A. et al., 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res., 39, D225-229.
Nacke H. et al., 2011. Identification of novel lipolytic genes and gene families by screening of metagenomic libraries derived from soil samples of the German Biodiversity Exploratories. FEMS Microbiol. Ecol., 78(1), 188-201.
Panda T. & Gowrishankar B.S., 2005. Production and applications of esterases. Appl. Microbiol. Biotechnol., 67(2), 160-169.
Petersen T.N. et al., 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8(10), 785-786.
Rao L. et al., 2011. A thermostable esterase from Thermoanaerobacter tengcongensis opening up a new family of bacterial lipolytic enzymes. Biochim. Biophys. Acta, 1814, 1695-1702.
Strohl W.R., 2000. The role of natural products in a modern drug discovery program. Drug Discovery Today, 5(2), 39-41.
Stroobants A. et al., 2014a. Diversity of bacterial communities in a profile of a winter wheat field: known and unknown members. Microb. Ecol., 68(4), 822-833.
Stroobants A. et al., 2014b. New carbohydrate-active enzymes identified by screening two metagenomic libraries derived from the soil of a winter wheat field. J. Appl. Microbiol., 117(4), 1045-1055.





